It seems like there has been a lot of cool wound-healing research results published lately. At the end of March, there was the result that bacteriophages hide their bacterial hosts from being cleared by mammalian immune response. Last week there was the result that aphids soldier nymphs use their own wound-healing proteins to plug gall holes. And now there's this new result on the mechanism underlying the role of long, noncoding RNA (lncRNA) in wound healing of human skin.
So what's long, non-coding RNA? The DNA in the human genome has to be converted to RNA, in much the same way computer code might be converted into assembly language, before it can then be converted into proteins (like assembly language being converted into object code that can actually be executed by the computer). Up until recently, biology has focused entirely on this pipeline -- DNA to RNA to protein. However, there are some bits of DNA that stop at RNA, which are termed "non-coding RNA" because they don't end up producing any proteins. They just float around until they naturally break down. However, there are hints that they are not merely intermediate transitional objects and that they can be functional molecules themselves. One of those hints is that they can be consistently turned on ("up-regulated") in certain situations. However, this doesn't mean that they're actually doing anything. There are technical details that make it useful to distinguish whether these non-coding RNA are short or long. I won't get into those. Just know, for the reset of this post, that "lncRNA" represents "long, non-coding RNA", which is a type of these mysterious molecules.
When healthy human skin is wounded, the normal response happens to involve the up-regulation of one of these lncRNA's is consistently generated more than in undamaged tissue, but no one knows why.
In this paper, they show that the molecule actually sequesters other mechanisms within the tissue that would normally turn off the migration of cells important for wound healing. So the lncRNA's are functional -- they get turned on when the skin is wounded, and that sets of a chain reaction that disables other mechanisms that would normally keep wound healing processes turned off. So the lncRNA's are an important step in the regulatory network. That's very cool. It not only helps scientists understand wound healing but also one more way in which lncRNA's can be functional -- not just junk molecules floating around waiting to be broken back down.
Here is the primary source:
"Human skin long noncoding RNA WAKMAR1 regulates wound healing by enhancing keratinocyte migration"
by Li et al.
PNAS (2019), early edition
https://doi.org/10.1073/pnas.1814097116
Significance
=====
Although constituting the majority of the transcriptional output of the human genome, the functional importance of long noncoding RNAs (lncRNAs) has only recently been recognized. The role of lncRNAs in wound healing is virtually unknown. Our study focused on a skin-specific lncRNA, termed “wound and keratinocyte migration-associated lncRNA 1” (WAKMAR1), which is down-regulated in wound-edge keratinocytes of human chronic nonhealing wounds compared with normal wounds under reepithelialization. We identified WAKMAR1 as being critical for keratinocyte migration and its deficiency as impairing wound reepithelialization. Mechanistically, WAKMAR1 interacts with DNA methyltransferases and interferes with the promoter methylation of the E2F1 gene, which is a key transcription factor controlling a network of migratory genes. This line of evidence demonstrates that lncRNAs play an essential role in human skin wound healing.
=====
Abstract
=====
An increasing number of studies reveal the importance of long noncoding RNAs (lncRNAs) in gene expression control underlying many physiological and pathological processes. However, their role in skin wound healing remains poorly understood. Our study focused on a skin-specific lncRNA, LOC105372576, whose expression was increased during physiological wound healing. In human nonhealing wounds, however, its level was significantly lower compared with normal wounds under reepithelialization. We characterized LOC105372576 as a nuclear-localized, RNAPII-transcribed, and polyadenylated lncRNA. In keratinocytes, its expression was induced by TGF-β signaling. Knockdown of LOC105372576 and activation of its endogenous transcription, respectively, reduced and increased the motility of keratinocytes and reepithelialization of human ex vivo skin wounds. Therefore, LOC105372576 was termed “wound and keratinocyte migration-associated lncRNA 1” (WAKMAR1). Further study revealed that WAKMAR1 regulated a network of protein-coding genes important for cell migration, most of which were under the control of transcription factor E2F1. Mechanistically, WAKMAR1 enhanced E2F1 expression by interfering with E2F1 promoter methylation through the sequestration of DNA methyltransferases. Collectively, we have identified a lncRNA important for keratinocyte migration, whose deficiency may be involved in the pathogenesis of chronic wounds.
=====
Personal weblog of Ted Pavlic. Includes lots of MATLAB and LaTeX (computer typesetting) tips along with commentary on all things engineering and some things not. An endless effort to keep it on the simplex.
Wednesday, April 24, 2019
Saturday, April 20, 2019
Stochastic versus random: The difference is whether you're describing a model or a focal system
It seems like there is a lot of confusion in quantitative modeling circles over the relationship between "randomness" and "stochasticity." This is in large part because the success of stochastic models to make sense of the world around us has led to wide use of stochastic modeling, so much that "stochastic" has become a near synonym for "random." However, the two terms are not identical.
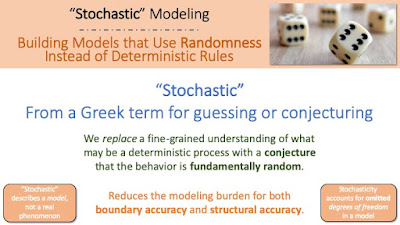 |
| Slide from SOS 212 (Systems, Dynamics, and Sustainability), an introductory modeling course I teach |
"Stochastic" is a term originating from Greek for "guess" or "conjecture." It describes a modeling technique to simplify a model by assuming (guessing/conjecturing) that the system being modeled is random, even if it is not. In other words, rather than having to account for a wide range of degrees of freedom that might be necessary to specify how a system is going to evolve, you substitute all of that detailed deterministic modeling with a crude approximation of the outcome being "random". The modeling burden then shifts from getting everything right down to high levels of precision to shaping the probability distributions to best match the outcomes that are most likely, regardless of what the underlying mechanism is. So a "stochastic model" is one that describes a system using randomness regardless of whether there is any reason to believe that the randomness is fundamental. It is a modeling trick to add analytically tractability to models that would otherwise be prohibitively complex to be useful.
So you shouldn't refer to phenomena themselves as being "stochastic." It is OK to say that the phenomena is random (if you really believe that). Or it is OK to say you use a stochastic model to describe the system (which implies that you will use randomness in order to omit a large number of degrees of freedom, thus making the model "simpler"). But you shouldn't use "stochastic" as a synonym for "random."
Wednesday, April 17, 2019
How to write a scientific paper, in four simple steps
Accomplished scientific writers will all have their own tips on how to go about writing a scientific paper. However, when you are first getting started, sometimes it is easier to have a set of steps available. Certainly there are many options available, but I personally have been finding the following set of steps useful when advising students lately.
- Step 1: Write the Introduction (starting with second paragraph)
The introduction lays out why you bothered to do the study you did. It provides a background from the existing literature, and it points out holes in that literature or opportunities to test answers to questions that have little coverage.
However, the first paragraph of your introduction (in fact, the first few sentences of your paper) are often the hardest ones to write. A colleague points out to her students that a great way to get around this potential writer's block is to simply skip the first paragraph. If you start with the second paragraph, you can immediately start laying out the argument for why your study needs to be done. Once you finish the rest of the introduction, it may be easy to see what the first paragraph needs to be. In other words, once you have 75% of the argument completed, the shape of the puzzle piece that remains will often be clear. - Step 2: Write the Discussion
If someone is already convinced that your paper has something interesting to say, that person will likely jump straight to the discussion and start reading there. This is where you describe how your data support some story that you want to tell about an inference you have made about the natural world. As you write your discussion, it will become clear exactly which data you need to tell your story. And so that brings us to the next step. - Step 3: Write the Results
Now that you have crafted a convincing discussion, you know what results are needed to support your argument. Writing the discussion first tells you what to include and what you can exclude. A lot of students make the mistake of starting with all of the possible plots that can be generated and then trying to string together a discussion around them, even if some of those plots really aren't needed. Don't do it that way. Instead, let your discussion guide which results to include. - Step 4: Write the Methods
Putting this step here is controversial. In many cases where your study has a clear experimental design that tests for something very specific, then it is possible to write your methods first. However, if you are instead working with a very large dataset that might someday result in multiple papers, then your methods might need to wait for you to determine exactly which results you need to make your argument. So once you've written your results, you can then generate methods that explain how all of those results came about.
Special Note: Title and Abstract
Of course, two things that are left out of the steps above are the title and abstract. Young writer's often leave these until the end, and they often view them as complementary to the rest of the article. However, with maturity, it should be possible to write them first and view them almost as alternatives (or marketing pieces) for the whole article. Writing them first can also provide structure (similar to an outline) for the rest of the article. So work toward being able to write these two things first, but it's OK if you start by writing them last.
Keep in mind that most readers will only read the title and abstract (and some will only read the title). Consequently, the title should not be clickbait; the title should be an executive summary of the article, giving the punchline as opposed to begging the reader to read further. Interested readers will move on to the abstract. Consequently, your abstract should really have everything an educated reader needs to reconstruct your article; it should have a little bit of all of the sections. Your abstract should concisely say why you did the things you did, how you did them, what the main results were, and why those results are interesting.
Take-away Message: It's About the ReaderOf course, two things that are left out of the steps above are the title and abstract. Young writer's often leave these until the end, and they often view them as complementary to the rest of the article. However, with maturity, it should be possible to write them first and view them almost as alternatives (or marketing pieces) for the whole article. Writing them first can also provide structure (similar to an outline) for the rest of the article. So work toward being able to write these two things first, but it's OK if you start by writing them last.
Keep in mind that most readers will only read the title and abstract (and some will only read the title). Consequently, the title should not be clickbait; the title should be an executive summary of the article, giving the punchline as opposed to begging the reader to read further. Interested readers will move on to the abstract. Consequently, your abstract should really have everything an educated reader needs to reconstruct your article; it should have a little bit of all of the sections. Your abstract should concisely say why you did the things you did, how you did them, what the main results were, and why those results are interesting.
People are busy, and they allocate their reading resources accordingly. If you tease them with half statements, they will only get frustrated and move on to the next thing that competes for their time. Instead, give them something interesting and complete in the title. If they want more details, they can read the abstract, which should itself be complete. If they want more details, they'll read the main body (possibly starting with the discussion section). For the reader, it's about sequentially choosing what to read next. When you write, you should always have this in mind as you design the delivery vehicle for the content you hope will be disseminated broadly.
Labels:
academia,
academic writing,
science,
scientific writing,
writer's block,
writing
Wednesday, April 10, 2019
Stochasticity, Randomness, and Chaos (and the differences between them)
In popular culture, words like "stochastic", "random", and "chaotic" are often used interchangeably. However, these three terms have totally different meanings. Furthermore, "randomness" and "chaos" are near opposites. Whereas "randomness" is used to simplify the process of model building, "chaos" is a phenomena that comes out of non-random models. Chaotic patterns appear random but are products of entirely deterministic processes. Chaos is an extreme sensitivity to initial conditions that relates to trajectories from deterministic systems gaining more and more individuality over time, as opposed to less. If it is chaotic, it is not random.
I explain the similarities and differences between stochasticity, randomness, and chaos in this two-part lecture recorded as a complement to material in my SOS 212 (Systems, Dynamics, and Sustainability) course at Arizona State University.
Lecture G1: Randomness and Chaos (SOS 212, Systems Dynamics and Sustainability, ASU)
For related videos that may be of interest, see my SOS 212 YouTube playlist.
I explain the similarities and differences between stochasticity, randomness, and chaos in this two-part lecture recorded as a complement to material in my SOS 212 (Systems, Dynamics, and Sustainability) course at Arizona State University.
Lecture G1: Randomness and Chaos (SOS 212, Systems Dynamics and Sustainability, ASU)
- Part 1: Randomness
- Part 2: Chaos
For related videos that may be of interest, see my SOS 212 YouTube playlist.
Bifurcation Diagrams, Hysteresis, and Tipping Points: Explanations Without Math
I teach a system dynamics modeling course (SOS 212: Systems, Dynamics, and Sustainability) at Arizona State University. It is a required course for our Sustainability BS students, which they ideally take in their second year after taking SOS 211, which is essentially Calculus I. The two courses together give them quantitative modeling fundamentals that they hopefully can make use of in other courses downstream and their careers in the future.
I end up having to cover a lot of content in SOS 212 that I myself learned through the lens of mathematics, but these students are learning it much earlier than I did and without many of the mathematical fundamentals. So I have to come up with explanations that do not rely on the mathematics. Here is an example from a recent lecture on bifurcation diagrams, hysteresis, and tipping points. It builds upon a fisheries example (from Morecroft's 2015 textbook) that uses a "Net regeneration" lookup table in lieu of a formal mathematical expression.
You can find additional videos related to this course at my SOS 212: System, Dynamics, and Sustainability playlist.
Subscribe to:
Posts (Atom)
